Study reveals evolutionary path of Influenza Viruses and its implications for vaccine development
Influenza A viruses, originating from avian sources, undergo crucial adaptations to human receptors for sustained transmission by targeting specific sialic acid configurations.
The recent study, published in the journal Cell Host & Microbe, has elucidated the evolutionary trajectory of H3N2 influenza viruses, providing insights into specific receptor interactions and antigenic variations crucial for vaccine development.
A collaborative international study, under the leadership of Professor James C. Paulson, from the Scripps Research Institute (La Jolla, USA) and including Jesús Jiménez-Barbero, Ikerbasque Professor and Scientific Director of CIC bioGUNE – member of BRTA – , have uncovered the secrets behind the flu’s ability to adapt and spread, offering valuable insights into vaccine development and public health strategies.
The recent article, published in the highly ranked scientific journal Cell Host & Microbe, offers a glimpse into the evolutionary journey of influenza viruses, with a special focus on the enduring H3N2 strain, which has persisted for over five decades. The study unveils how these viruses have evolved to target specific receptors on human airway cells, providing essential knowledge for vaccine development and understanding virus transmission dynamics.
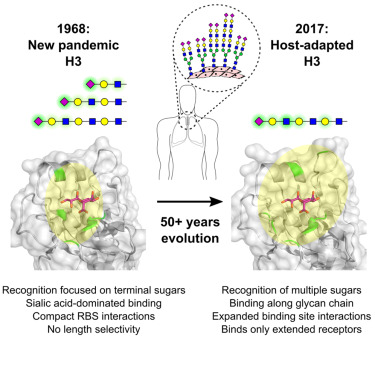
Influenza A viruses (IAVs) have historically caused significant respiratory illness, including seasonal outbreaks and global pandemics. These viruses originate from avian sources and occasionally acquire the ability to infect humans. IAV strains are named based on their surface proteins, HA and NA, which interact with receptors on airway cells. Human IAVs typically target receptors with specific sialic acid configurations, while avian viruses have different preferences. Adaptation of avian HAs to human receptors is crucial for sustained transmission in humans.
Pandemic IAV strains, like H1, H2, and H3, undergo mutations to shift receptor preference. H3 viruses have evolved to recognize more complex receptor structures over time, involving mutations outside the traditional receptor-binding site. The research described here emphasizes the intricate relationship between HA mutations and receptor binding, with even minor changes requiring compensatory mutations.
The study focuses on H3N2 viruses, highlighting their continuous adaptation over five decades of circulation in the human population and also disentangling the fine details of the molecular requirements to fit such adaptation using a multidisciplinary scientific approach, which have involved members of Scripps, CIC bioGUNE, CIB-CSIC, Universidad Complutense (Madrid) and CIBERES.
The results obtained shed light on how flu viruses infect cells by targeting specific molecules on airway cells, aiding researchers in understanding transmission dynamics and the varying severity of strains. It underscores the need for updated vaccines as flu viruses evolve, ensuring better protection for the public. Understanding virus evolution enables better preparedness for future outbreaks, empowering public health officials to take proactive measures.